Prediction Intervals with specific value prediction
This is an implementation of the model described in the following paper:
Simhayev, Eli, Gilad Katz, and Lior Rokach. “PIVEN: A Deep Neural Network for Prediction Intervals with Specific Value Prediction.” arXiv preprint arXiv:2006.05139 (2020).
I have copied some of the code from the paper’s code base, and cite the author’s paper where this is the case.
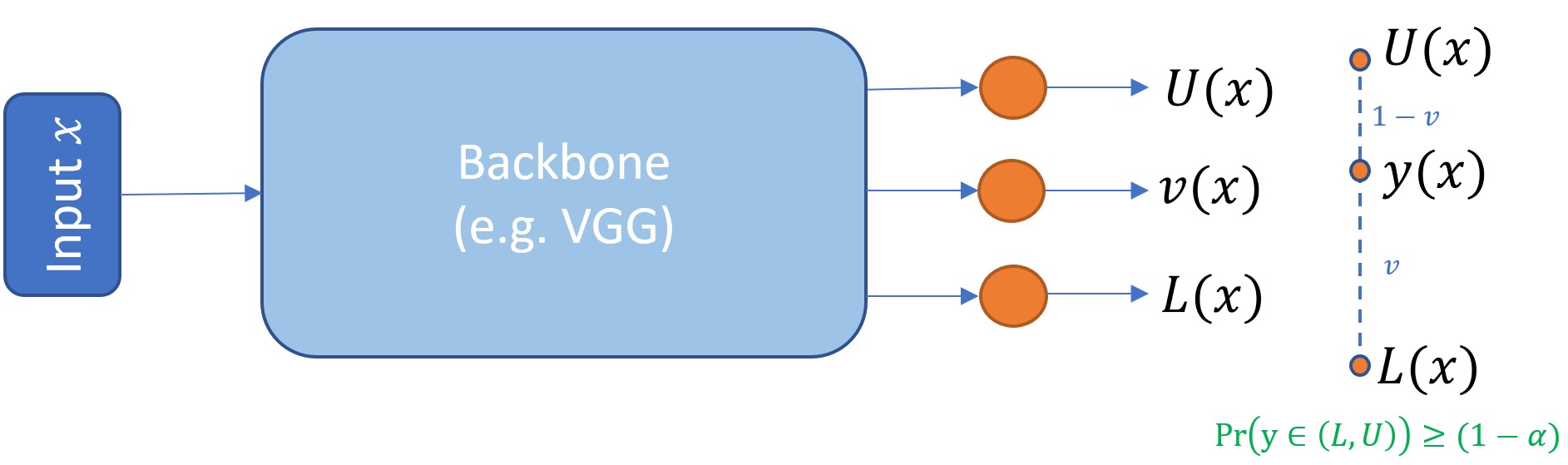
A neural network with a Piven (Prediction Intervals with specific value prediction) output layer returns a point
prediction as well as a lower and upper prediction interval (PI) for each target in a regression problem.
This is useful because it allows you to quantify uncertainty in the point-predictions.
Using the piven module is quite straightforward. For a simple MLP with a piven output, you can run:
import numpy as npfrom piven.models import PivenMlpModelfrom sklearn.preprocessing import StandardScaler# Make some dataseed = 26783np.random.seed(seed)# create some datan_samples = 500x = np.random.uniform(low=-2.0, high=2.0, size=(n_samples, 1))y = 1.5 * np.sin(np.pi * x[:, 0]) + np.random.normal(loc=0.0, scale=1 * np.power(x[:, 0], 2))x_train = x[:400, :].reshape(-1, 1)y_train = y[:400]x_valid = x[400:, :].reshape(-1, 1)y_valid = y[400:]# Build piven modelmodel = PivenMlpModel(input_dim=1,dense_units=(64, 64),dropout_rate=(0.0, 0.0),lambda_=25.0,bias_init_low=-3.0,bias_init_high=3.0,lr=0.0001,)# Normalize input datamodel.build(preprocess=StandardScaler())# You can pass any arguments that you would also pass to a keras modelmodel.fit(x_train, y_train, model__epochs=200, model__validation_split=.2)
The image below shows how the lower and upper PI change as we keep training the model

You can score the model by calling the score() method:
y_pred, y_ci_low, y_ci_high = model.predict(x_test, return_prediction_intervals=True)model.score(y_true, y_pred, y_ci_low, y_ci_high)
To persist the model on disk, call the save() method:
model.save("path-to-model-folder", model=True, predictions=True)
This will save the metrics, keras model, and model predictions to the folder.
If you want to load the model from disk, you need to pass the model build function (see below for more information).
from piven.models import piven_mlp_modelmodel = PivenMlpModel.load("path-to-model-folder", build_fn=piven_mlp_model)
For additional examples, see the ‘tests’ and ‘notebooks’ folders.
You can use a Piven layer on any neural network architecture. The authors of the Piven paper use it on top of
a pre-trained CNN to predict people’s age.
Suppose that you want to create an Model with a Piven output layer. Because this module uses the
KerasRegressor wrapper
from the tensorflow library to make scikit-compatible keras models, you would first specify a build
function like so:
import tensorflow as tffrom piven.layers import Pivenfrom piven.metrics.tensorflow import picp, mpiwfrom piven.loss import piven_lossdef piven_model(input_size, hidden_units):i = tf.keras.layers.Input((input_size,))x = tf.keras.layers.Dense(hidden_units)(i)o = Piven()(x)model = tf.keras.models.Model(inputs=i, outputs=o)model.compile(optimizer="rmsprop", metrics=[picp, mpiw],loss=piven_loss(lambda_in=15.0, soften=160.0,alpha=0.05))return model
The most straightforward way of running your Model is to subclass the PivenBaseModel class. This requires you
to define a build() method in which you can add preprocessing pipelines etc.
from piven.models.base import PivenBaseModelfrom piven.scikit_learn.wrappers import PivenKerasRegressorfrom piven.scikit_learn.compose import PivenTransformedTargetRegressorfrom sklearn.preprocessing import StandardScalerclass MyPivenModel(PivenBaseModel):def build(self, build_fn = piven_model):model = PivenKerasRegressor(build_fn=build_fn, **self.params)# Finally, normalize the output targetself.model = PivenTransformedTargetRegressor(regressor=model, transformer=StandardScaler())return self
To initialize the model, call:
MyPivenModel(input_size=3,hidden_units=32)
Note that the inputs to MyPivenModel must match the inputs to the piven_model function.
You can now call all methods defined as in the PivenBaseModel class. Check the
PivenMlpModel class
for a more detailed example.
The piven loss function is more complicated than a regular loss function in that it combines three objectives:
The piven loss function combines these objectives into a single loss. The loss function takes three arguments.
The default settings are those used by the authors of the paper. You should probably leave them as they are unless you
know what you are doing. For further details, see [1, pp. 4-5].
In statistics/ML, uncertainty is often subdivided into ‘aleatoric’ and ‘epistemic’ uncertainty. The former is associated
with randomness in the sense that any experiment that is not deterministic shows variability in its outcomes. The latter
type is associated with a lack of knowledge about the best model. Unlike aleatoric uncertainty, epistemic uncertainty
can be reduced by acquiring more information. [2].
Prediction intervals are always wider than confidence intervals, since confidence intervals try to capture epistemic
uncertainty only whereas prediction intervals seek to capture both types. See pages 2 and 5 in [1] for a discussion
on quantifying uncertainty.
[1] Simhayev, Eli, Gilad Katz, and Lior Rokach. “PIVEN: A Deep Neural Network for Prediction Intervals with Specific Value Prediction.” arXiv preprint arXiv:2006.05139 (2020).
[2] Hüllermeier, Eyke, and Willem Waegeman. “Aleatoric and epistemic uncertainty in machine learning: A tutorial introduction.” arXiv preprint arXiv:1910.09457 (2019).